Hello! I am Parmida Ghahremani. Computer Vision and Deep Learning Research Scientist.
I am a senior data scientist at Memorial Sloan Kettering Cancer Center. I am working on Computer Vision and Deep Learning as well as data synthesis and collection techniques for segmentation, registration, classification and visualization of objects in biomedical images.

Paper GitHub Repository DeepLIIF Website
- Designed and implemented a GAN-Based multitask deep learning model for single-step translation, segmentation, and classification. Achieved this by designing a universal optimizer and weighted loss function, outperforming state-of-the-art models such as Mask-RCNN and nnUNet on segmentation and classification tasks.
- Designed and implemented a GAN-based image synthesis model for generating high fidelity IHC images using corresponding Hematoxylin and Marker images by defining style- and feature-based loss function.
- Developed a framework offering rigid transformations for co-registering IHC and mpIF data using Tkinter python interface.
- Implemented computer vision algorithms for pre- and post-processing data using skimage, scipy, numba, and matplotlib.

- Developed an end-to-end application (NeuroConstruct) for reconstruction and visualization of 3D neuronal structures.
- Designed and implemented a novel 3D nested UNet-based network with skip pathways for segmenting objects in volumes using Tensorflow and Keras, outperforming state-of-the-art models including U2Net, UNet++, and UNet3+.
- Designed a hybrid rendering approach, combining iso-surface rendering of high-confidence classified neurites, along with real-time rendering of raw volume.
- Created segmentation and registration toolbox for auto \& manual segmentation of neurons and coarse-to-fine alignment of serial brain sections with 3D rendered volume, 2D cross-sectional views, and novel annotation functions using PyQt5 and vtk.

- Created a semi-automatic crowdsourcing framework for nuclei segmentation in pathology slides, allowing publication of jobs containing question and judgment phases on Amazon Mechanical Turk for collecting ground-truth segmentation dataset.
- Designed and implemented a novel CNN approach (CrowdDeep) for nuclei segmentation using a combination of crowd and expert annotations, outperforming expert-trained-only models.
- Developed a visual analytic framework for evaluation of CrowdDeep using D3 visualization.

- Conducted two VR user studies to evaluate our techniques over a search and comparison task using Unity Game Engine, HTC Vive headset, and controllers.
- Performed quantitave analysis in R to evaluate our novel approaches against state-of-the-art methods using traditional and proposed questionnaires and metrics including SSQ, NASA-TLX, Presence, Performance, Tapping Test, and Movement with two-way RANOVA and Tukey's ladder of power.

Unsupervised Co-segmentation of 3D shapes via clustering
Stony Brook University, CSE 528 (C++, OpenGL)
- Co-segmenting and visualization of 3D shapes via subspace clustering by partitioning each shape into primitive patches using normalized cuts (NCuts) and computing feature descriptors for each patch.
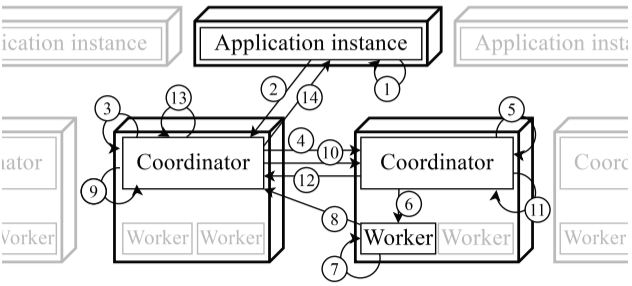
Scalable and secure concurrent evaluation of history-based access control policies
Stony Brook University, CSE 535 (Python, DistAlgo)
- Developing the distributed coordinator proposed in ”Scalable and Secure Concurrent Evaluation of History-based Access Control Policies”.
- Extended it to a multi-version concurrency control algorithm with timestamp ordering in DistAlgo.
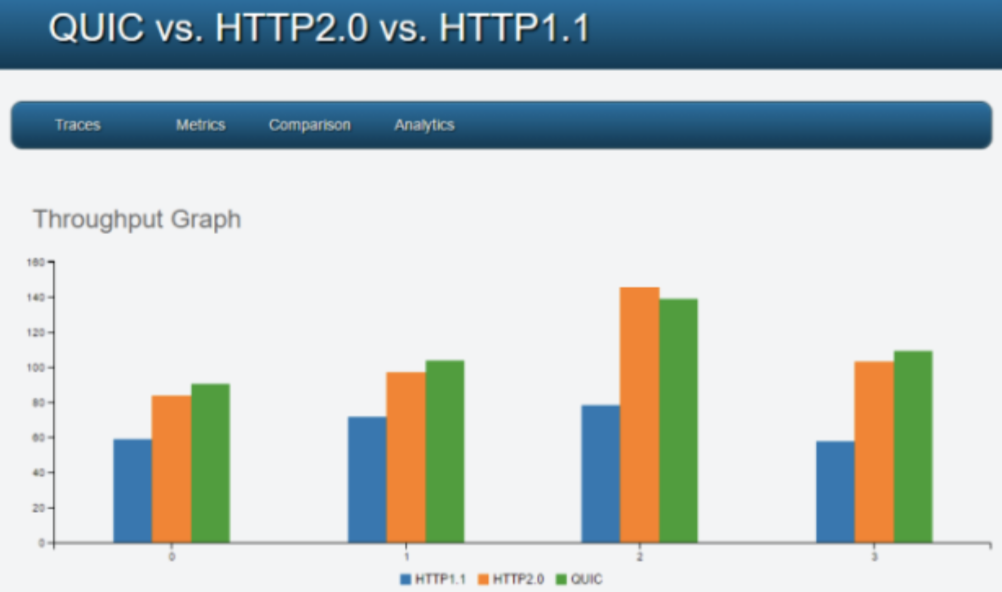
Visualization tool for network packets analysis
Stony Brook University, CSE 564 (Python, D3 visualization, Wireshark)
- Designing a visualization tool to analyze and compare characteristics of SPDY and HTTP packets.
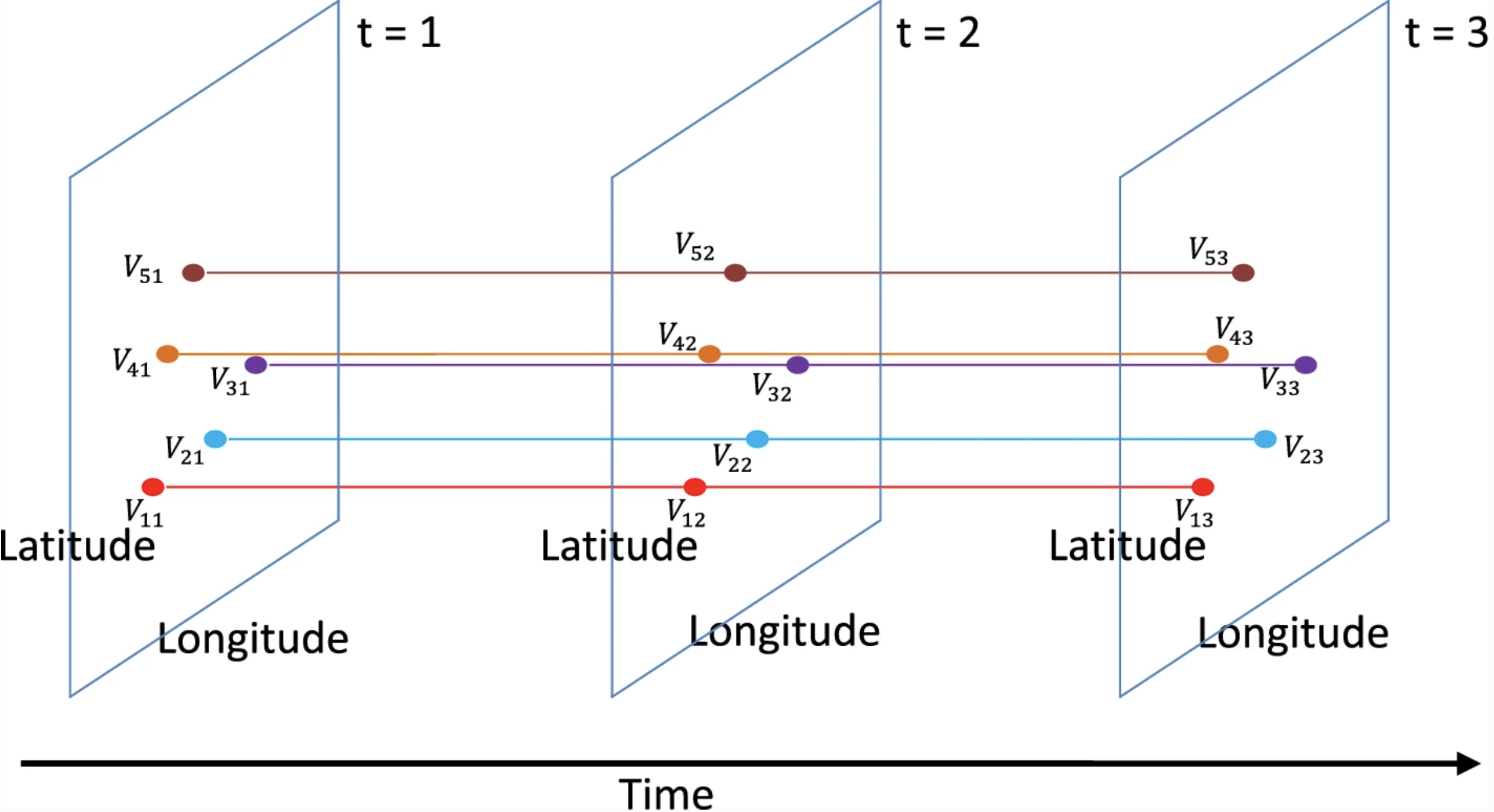
On-line aggregation for interactive analysis over large-scale spatial and temporal data
Stony Brook University, CSE 532 (Java, Hadoop)
- Implementing a temporal and spatial SQL query executor system, supporting all spatial operators.
February 2022 - Present
Designing deep learning approaches for various medical image analysis tasks.
May 2021 - September 2021
Designed deep learning approaches for cell and membrane segmentation in IHC and fluorescence microscopy images.
June 2015 - August 2015
Designing an interactive offline map with informative pins on locations.
2017 - 2022
Stony Brook University
DeepLIIF: Deep-Learning Inferred Multiplex Immunofluoresence for IHC Image Quantification
Python, PyTorch
By creating a multitask deep learning framework called DeepLIIF, we present a single-step solution to stain deconvolution/separation, cell segmentation, and quantitative single-cell IHC scoring. Leveraging a unique de novo dataset of co-registered IHC and multiplex immunofluorescence (mpIF) staining of the same slides, we segment and translate low-cost and prevalent IHC slides to more expensive-yet-informative mpIF images, while simultaneously providing the essential ground truth for the superimposed brightfield IHC channels.
NeuroConstruct: 3D Reconstruction and Visualization of Neurites in Optical Microscopy Brain Images
Python, C++, Keras, Tensorflow, FluoRender, ImageJ, Matlab
In this project, we reconstruct and visualize 3D neuronal structures in wide-field microscopic images. NeuroConstruct offers a Segmentation Toolbox to precisely annotate micrometer resolution neurites. It also offers an automatic neurites segmentation using 2D and 3D CNNs trained by the Toolbox annotations. To visualize neurites in a given volume, NeuroConstruct offers a hybrid rendering by combining iso-surface rendering of high-confidence classified neurites, along with real-time rendering of raw volume. It also introduces a Registration Toolbox for automatic coarse-to-fine alignment of serially sectioned samples.
CrowdDeep: nuclei detection and segmentation using crowdsourcing and deep learning
Python, Keras, Tensorflow, Amazon Mechanical Turk, JavaScript (D3 visualization)
In this project, we designed a crowdsourcing framework for nuclei segmentation in pathology slides, and after tiling the slides, published tiles on Amazon Mechanical Turk to be annotated by the crowd. Then, the crowd annotated images are used for training a convolutional neural network to detect and segment nuclei in pathology slides.
2015 - 2017
Stony Brook University
Exploration of Large Omnidirectional Images in Immersive Environments
C#, UnityR Game Engine, R
We focused on visualizing and navigating large omnidirectional or panoramic images with application to GIS visualization as an inside-out omnidirectional image of the earth using UnityR Game Engine, HTC Vive headset and controllers. Then, we conducted two user studies involving 40 people and 185 individual cases, to evaluate our techniques over a search and comparison task. Our results illustrate the advantages of our techniques for navigation and exploration of omnidirectional images in an immersive environment such as less mental load and greater flexibility.
2011 - 2015
Sharif University of Technology
B.Sc. Thesis: Workload characterization of buffer cache layer in Linux operating system
In this work, we proposed an efficient data migration scheme at the Operating System level in a hybrid DRAM-NVM memory architecture by preventing unnecessary migrations and only allowing migrations with benefits to the system in terms of power and performance. The experimental results show that the proposed scheme reduces the hit ratio in NVM and improves the endurance of NVM resulting in significantly higher performance and less power consumption.
Cancer Simulation: We designed a system for simulation of DCIS Cancer cells growth by implementing an agent-based model of tumor growth driving from Macklin’s model, followed by application of evolutionary game theory (EGT) to model the interactions between adjacent cancer cells via gap junctions. This system was implented in Java and used in the paper ”Integrating Evolutionary Game Theory into an Agent-Based Model of Ductal Carcinoma in Situ: Role of Gap Junctions in Cancer Progression” published in Computer Methods and Programs in Biomedicine.
